Abstract
Multi-modal medical image registration plays a critical role in neuroimaging applications, enabling spatial alignment between modalities such as CT and MRI. In this study, we conduct a comparative analysis of classical and deep learning-based registration algorithms on the RIRE dataset. We evaluate rigid, affine, and deformable classical methods alongside deep models, including BrainMorph, LapIRN, and uniGradICON, using SSIM, MSE, and visual assessments.
Our results demonstrate that deep learning methods generally outperform classical algorithms in terms of accuracy and robustness, but classical techniques remain competitive in runtime and stability under rigid transformations.
1. Introduction
Multi-modal medical image registration is essential for aligning anatomical structures across imaging modalities like CT and MRI. Classical methods optimize similarity metrics such as mutual information (MI) or normalized cross-correlation (NCC), but struggle under large intensity variations or non-rigid structures. Deep learning methods, both supervised and unsupervised, now offer powerful data-driven alternatives.
However, unified benchmarks across both paradigms are scarce. This study aims to bridge that gap by evaluating classical and learning-based registration methods on CT-MRI brain images from the RIRE dataset.
2. Related Work
2.1 Classical Image Registration
Classical techniques optimize similarity metrics over transformation models:
- Mutual Information (MI): Effective for multi-modal settings (Maes et al., 1997).
- Normalized Cross-Correlation (NCC): Common in intra-modality registration.
- FFT-based Registration: Fast but limited to translations (Reddy & Chatterji, 1996).
- Feature-Based (RANSAC-SIFT): Robust to modality differences but reliant on texture quality.
2.2 Deep Learning for Registration
Deep learning revolutionized registration:
- VoxelMorph: Unsupervised deformation prediction (Balakrishnan et al., 2019).
- LapIRN: Coarse-to-fine hierarchical refinement (Mok & Chung, 2020).
- uniGradICON: Universal foundation model (Tian et al., 2024).
- BrainMorph: Supervised keypoint-based registration (Wang et al., 2024).
2.3 Clinical Relevance
Clinical deployment demands not only high SSIM but robust, anatomically plausible deformations. Existing evaluations often neglect real-world multi-modal challenges.
3. Implementation
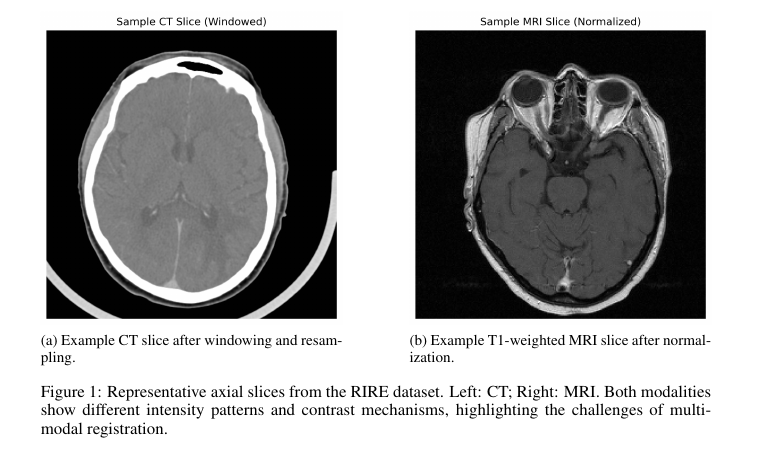
3.1 Dataset and Preprocessing
- Dataset: RIRE (14 patients, CT and T1-weighted MRI).
- Preprocessing:
- CT windowed to [-160, 240] HU and normalized.
- MRI volumes linearly normalized.
- Deep models processed full 3D volumes; classical methods on 2D central slices.
3.2 Classical Methods
- MI and NCC rigid/affine alignment using SimpleITK.
- FFT Phase Correlation for translation-only shifts.
- Deformable Registration with Gaussian smoothing.
- RANSAC-SIFT keypoint-based affine matching.
3.3 Deep Learning Methods
- BrainMorph: Supervised, keypoint detection, TPS deformation.
- LapIRN: Unsupervised, coarse-to-fine Laplacian refinement.
- uniGradICON: Universal model, invertibility-regularized U-Nets.
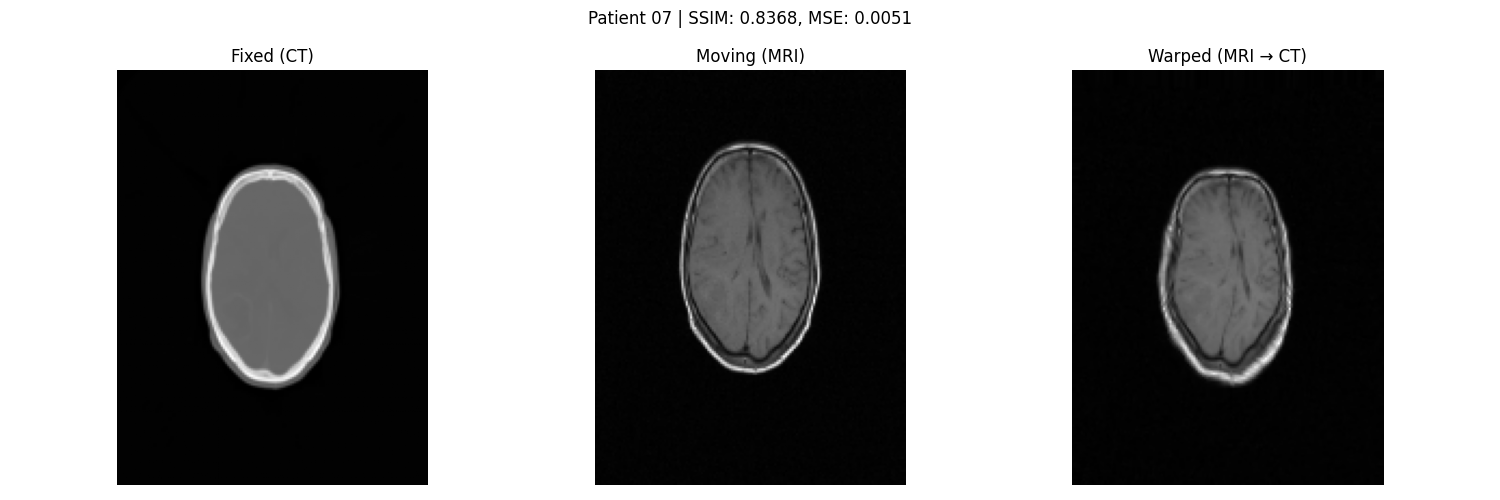
3.4 Evaluation Metrics
- SSIM and MSE between registered MRI and CT.
- Runtime profiling.
- Visual assessment through overlays.
4. Experiments
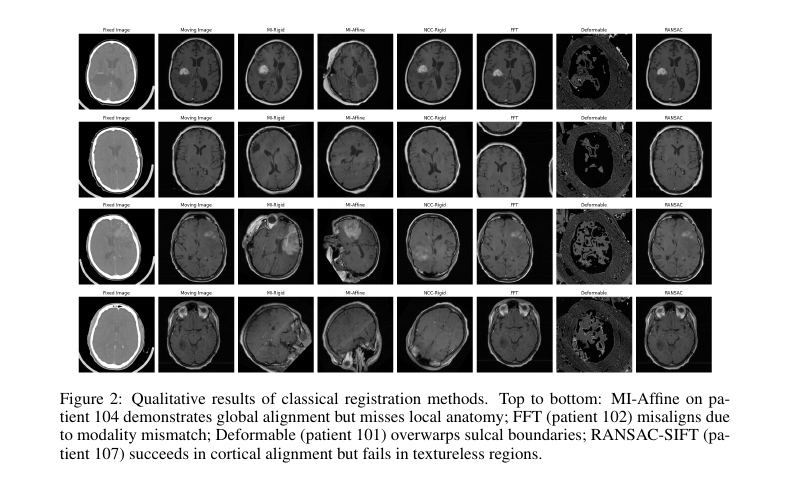
4.1 RQ1: Anatomical Fidelity and Failure Modes
- Classical methods:
- MI-Affine achieves gross alignment but lacks local deformation.
- FFT fails under intensity shifts.
- Deformable overwarps.
- RANSAC-SIFT is sensitive to texture.
- Deep models:
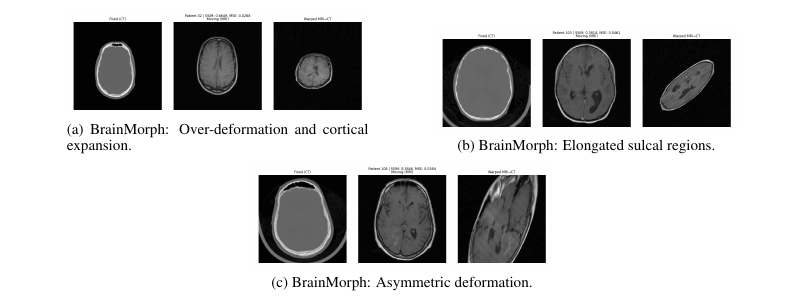
- BrainMorph: Accurate where keypoints are rich; over-deforms elsewhere.
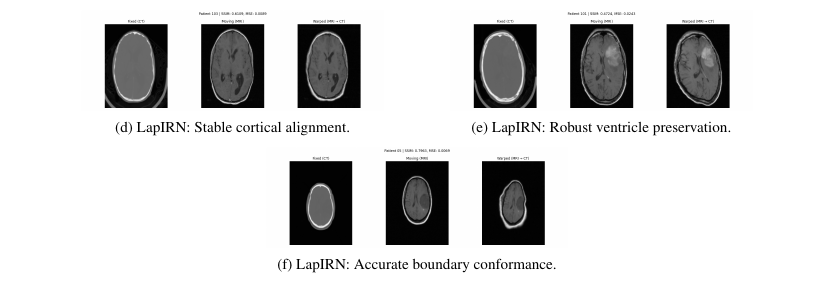
- LapIRN: Best balance of precision and smoothness.
- uniGradICON: Excellent global smoothness but slightly less sharp at fine detail.
![]()
4.2 RQ2: Accuracy vs Computational Cost
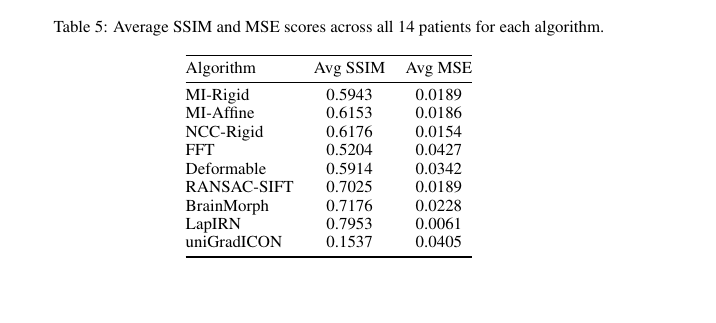
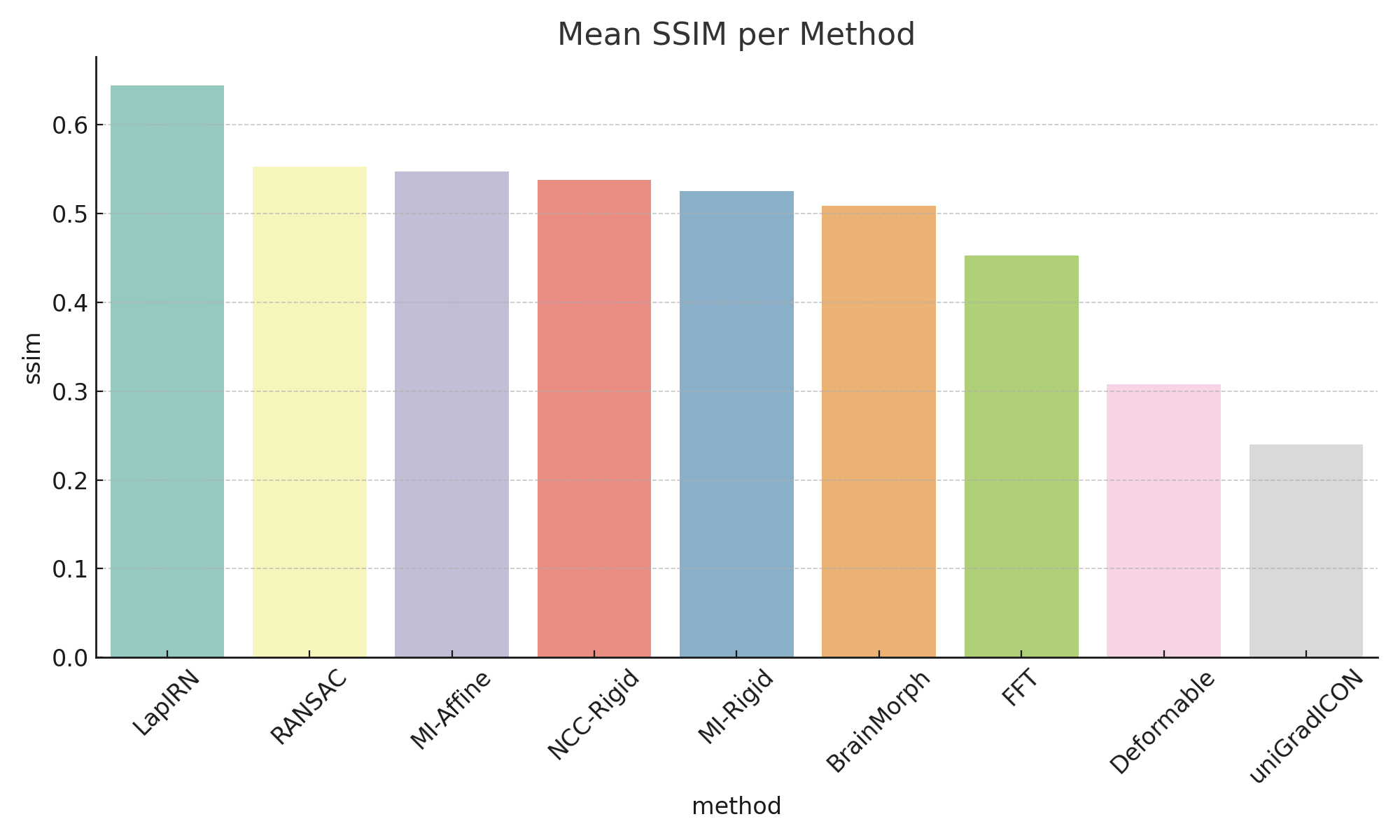
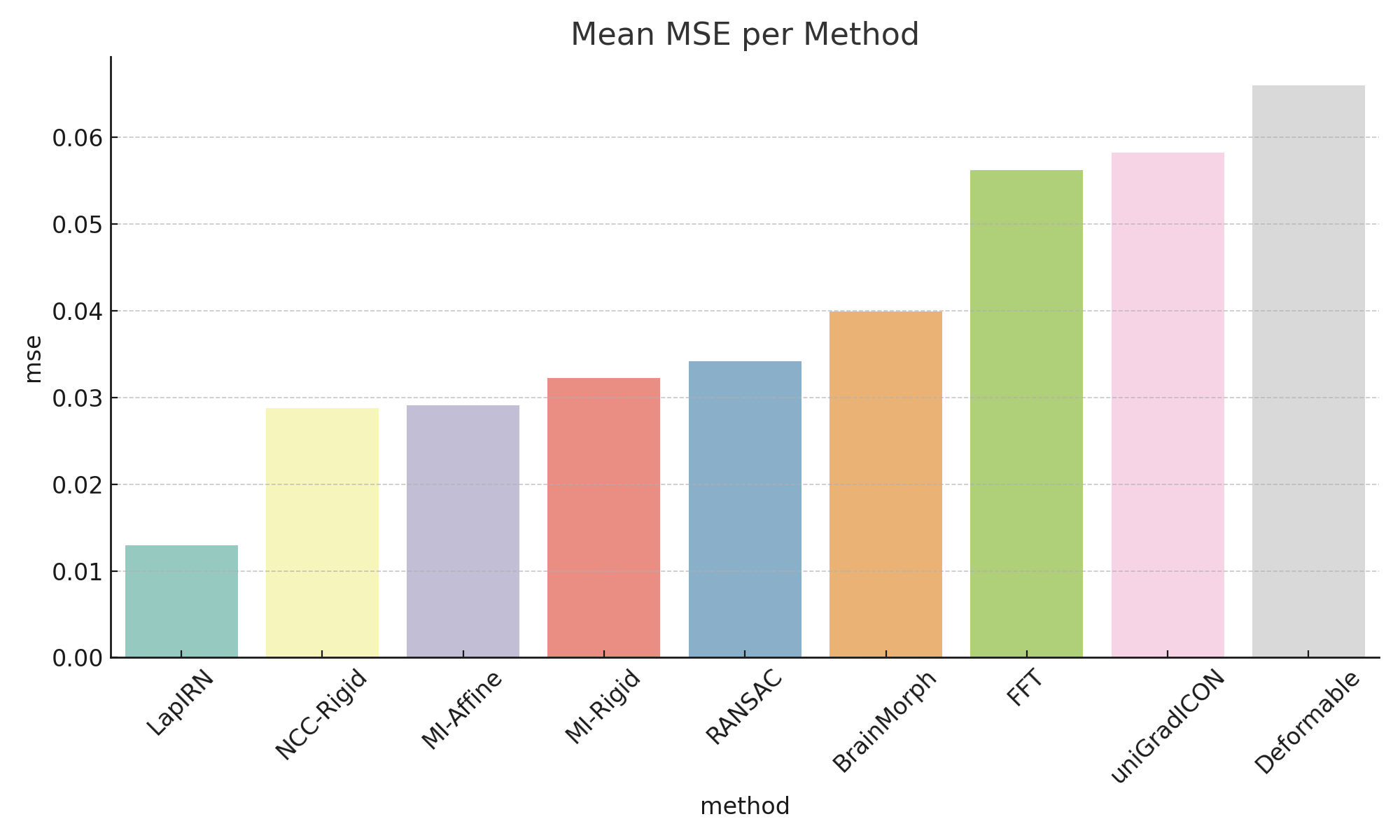
- Deep models achieve higher SSIM and lower MSE.
- Classical methods are much faster but less accurate.
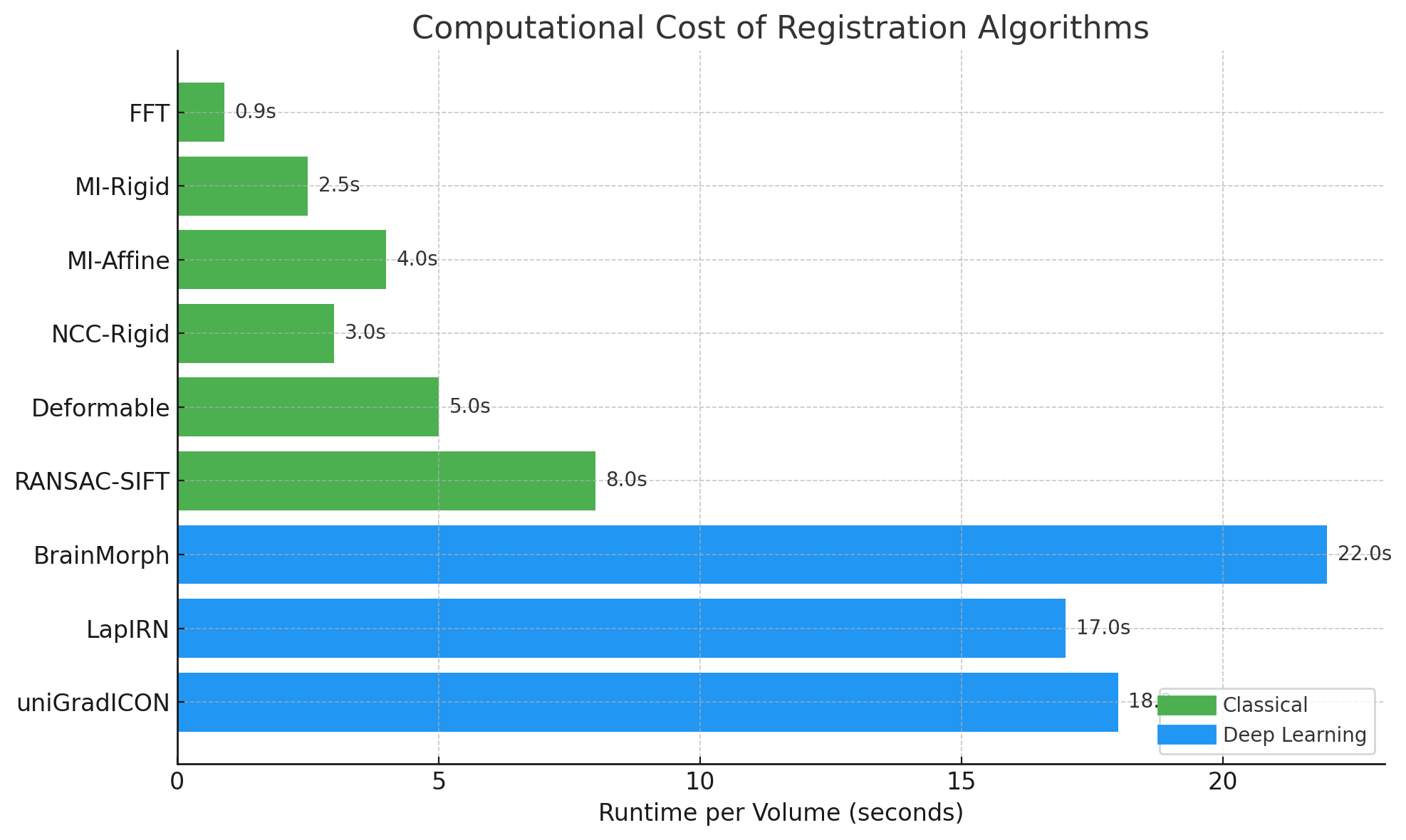
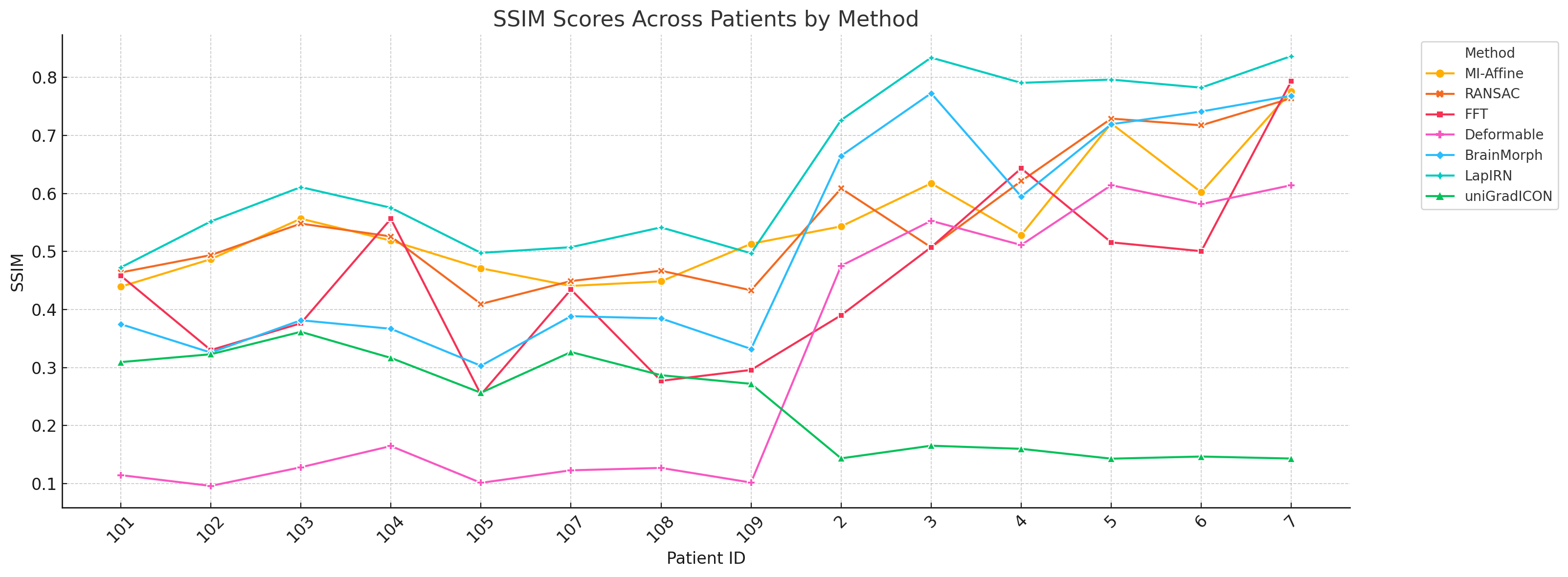
4.3 RQ3: Consistency Across Patients
- LapIRN: Most consistent performance across anatomical variability.
- BrainMorph: Sensitive to keypoint density.
- Classical methods: Highly variable results across patients.
5. Limitations
- Small dataset (14 patients).
- Pretrained deep models without fine-tuning.
- Classical methods evaluated only on 2D slices.
- SSIM and MSE don’t fully capture clinical registration quality.
- Runtime measured on a specific hardware setting (Google Colab Pro).
6. Conclusion
Deep learning models significantly outperform classical methods in multi-modal brain image registration, especially in deformable cases.
- LapIRN offers the best balance of accuracy, smoothness, and robustness.
- BrainMorph excels where keypoints are dense but can over-deform.
- uniGradICON generalizes broadly but trades off fine detail.
Classical methods remain viable for fast, rigid registration under constrained settings.
References
- Balakrishnan et al., “VoxelMorph”, IEEE Transactions on Medical Imaging, 2019. DOI
- Mok & Chung, “LapIRN”, 2020. arXiv:2006.16148
- Tian et al., “uniGradICON”, 2024. arXiv:2403.05780
- Wang et al., “BrainMorph”, 2024. arXiv:2405.14019
- Maes et al., “Mutual Information for Registration”, IEEE TMI, 1997.
- Reddy & Chatterji, “FFT-based Image Registration”, IEEE TIP, 1996.